About: Brain Decoding Project
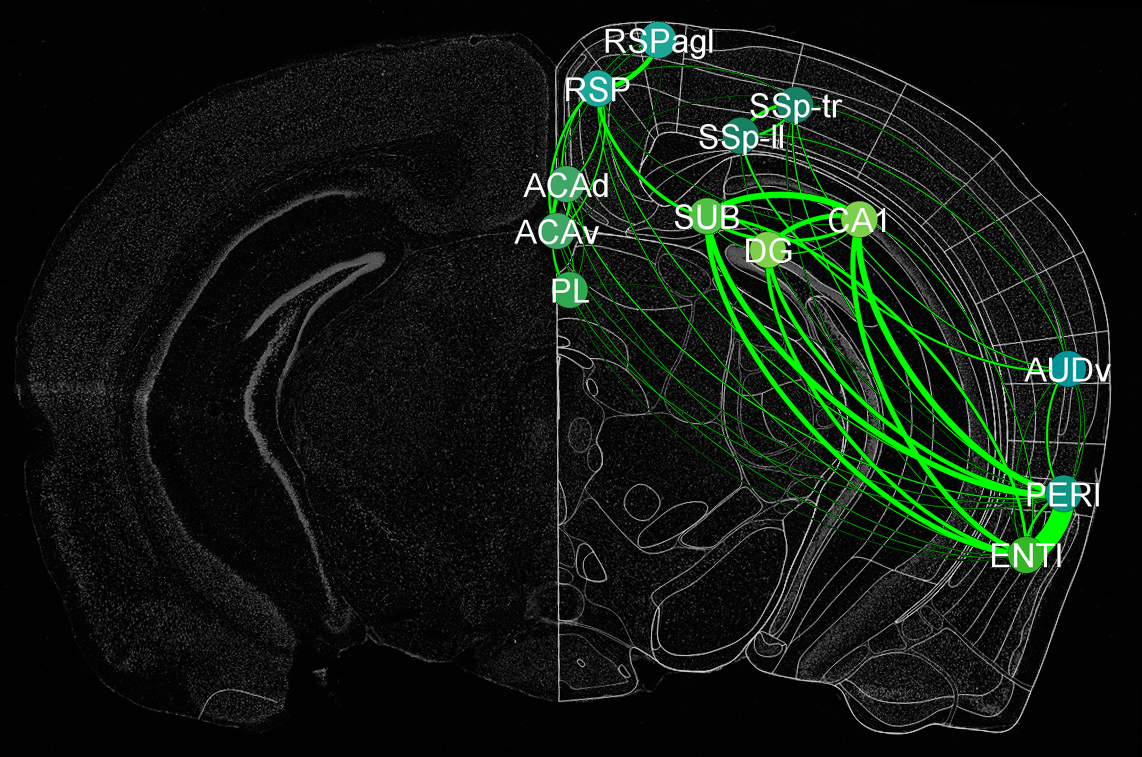
Cracking the neural code and neural circuit dynamics face unique sets of problems: 1) The ability to monitor large-scale neural activity patterns during cognitive behaviors; 2) lack of information on the component cell types and their functional contributions to neural coding; 3) require computational tools to investigate complex dynamic patterns of neural activity, thereby understanding real-time neural code; and 4) lack of fundamental knowledge about the organizing principles that govern real-time neural codes across various interacting circuits in the brain as a whole. In the learning and memory field, little is known about: What are real-time memory traces at the neural population level? How do short-term memory traces differ from long-term memory traces? How do distinct memory traces from multiple brain regions get synthesized into a holistic memory engram at the whole brain level?
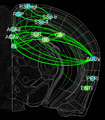
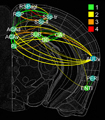
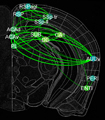
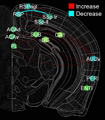
In this project, we will employ our newly developed large-scale electrodes arrays techniques to record neural activity patterns over a dozen brain sites simultaneously as mice undergo learning, consolidation, and memory recalls of emotionally charge experiences. The large-scale neural recording will be further accompanied with cell type-specific optogenetic manipulations to further reveal the underlying cell assembly architectures and their interaction across the broad memory-processing circuits. We will apply and develop innovative computational and mathematical approaches to uncover real-time memory codes and provide conceptual insights into the network-level organization of fear memory and associative working memory in the mouse brain. Finally, to facilitate the transformative, paradigm-shift in sharing large-scale neurophysiological datasets, we propose to create an open-access Mouse Memory Code Database (MMCD) platform for stable long-term data storage and dissemination to broad communities of researchers.
[link: Decoding the Brain]
[link: History of Brain Decoding Project]
 Dr. Joe Tsien (
Dr. Joe Tsien ( Dr. Tianming Liu is an associate professor in the Computer Science Department at the University of Georgia in Athens (
Dr. Tianming Liu is an associate professor in the Computer Science Department at the University of Georgia in Athens ( Dr. Guantao Chen is the Distinguished University Professor and Chair of the Department of Mathematics and Statistics at Georgia State University (
Dr. Guantao Chen is the Distinguished University Professor and Chair of the Department of Mathematics and Statistics at Georgia State University ( Dr. King-Ip (David) Lin is an associate professor at the Department of Computer Science of the University of Memphis and he is an expert in database research (
Dr. King-Ip (David) Lin is an associate professor at the Department of Computer Science of the University of Memphis and he is an expert in database research (